Dr. Ye Ai is currently an Associate Professor at Singapore University of Technology and Design (SUTD). He obtained his B.S. in Mechanical Engineering from Huazhong University of Science and Technology (China) in 2005 and his Ph.D. in Mechanical and Aerospace Engineering from Old Dominion University (USA) in 2011. Prior to joining SUTD as an assistant professor in 2013, he worked as a postdoctoral researcher at the Bioscience Division of Los Alamos National Laboratory from June 2011 to January 2013. He was a visiting scholar at Massachusetts Institute of Technology (MIT) from August 2014 to July 2015. He was promoted to associate professor with tenure in September 2019. Dr. Ai’s research interest focuses on developing novel microfluidic technologies for particle/cell manipulation and single cell analysis. His research team is also striving to translate their innovative microfluidic technologies to commercial market through collaborations with industry.
Read Dr Ai’s recent Emerging Investigator Series paper: Microfluidic impedance cytometry device with N-shaped electrodes for lateral position measurement of single cells/particles in the most recent issue of Lab on a Chip, and find out more about him and his work below.
- Your recent Emerging Investigator Series paper focuses on measuring the lateral position of single cells or particles. How has your research evolved from your first article to this most recent article?
My first research article when I was a PhD student was to develop a finite element model with dynamically deformed mesh that can simulate the transient motion of finite-size particles in microscale fluid flows. My PhD research mainly focused on electrokinetics for manipulating particles, cells and ions in micro/nanoscale. My postdoctoral training at the Bioscience Division of Los Alamos National Laboratory exposed me to a lot of biological problems, in particular the need in high-throughput cellular analysis at the single cell level. My previous research experience has somehow shaped my current research focus into single cell manipulation and analysis using novel microfluidic technologies when I become an independent principal investigator in Singapore.
- What aspect of your work are you most excited about at the moment?
I am most excited to apply our developed microfluidic technologies for solving real biomedical problems and enabling new biological studies. As an example, in 2017 my team published our single cell sorting technology using a highly focused acoustic beam in Lab Chip (DOI: 10.1039/c7lc00678k). Later, I was approached by quite a number of research teams worldwide who wanted to try our sorting technology. These email communications have encouraged me to apply our developed prototype for real biomedical problems. Right now, I have established collaboration with a few biomedical research institutes in Singapore and we have found that our sorting technology is not causing any cell damage, which is however challenging for conventional FACS machine. We currently have the idea to commercialize this single cell sorting technology. Let us see what is going to happen in the next few years.
- In your opinion, what are the key considerations when designing a microfluidic platform for real-time measurements?
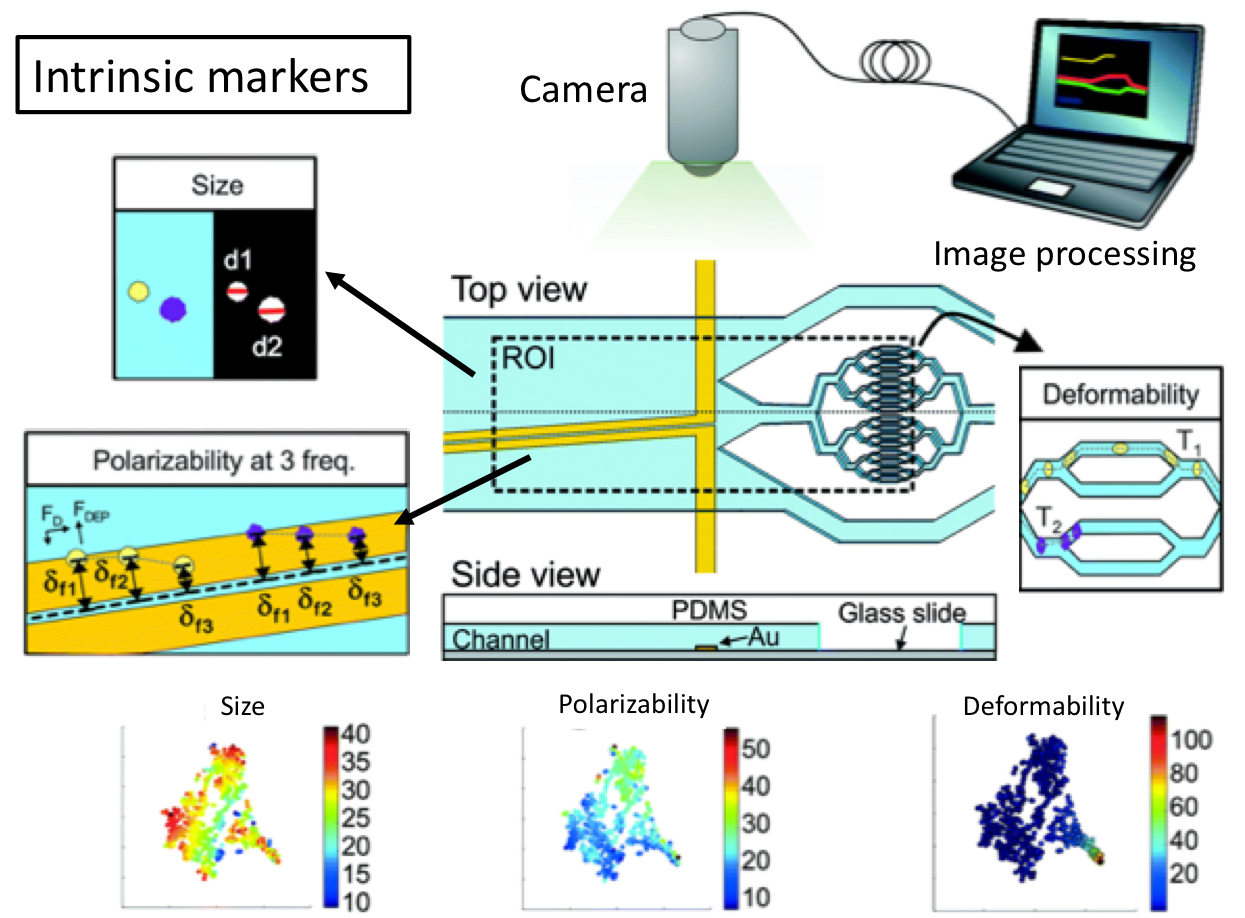
My research team is currently developing both hydrodynamic and acoustic cell sorting platforms. The conventional way to quantify the sorting performance (e.g. purity and recovery) is to run additional cell analysis of collected samples, typically using a flow cytometer. In this work (DOI: 10.1039/c9lc00819e), we designed and validated a new impedance cytometry device that enables the measurement of the lateral positions and physical properties of individual particles and cells. The integration of this new device with any cell sorting platform will allow the evaluation of the sorting performance to be implemented in the same system.
The key consideration of integrating these real-time measurements really depends on whether there is a critical need. But I do see a lot of sorting applications need these real-time, in-line measurements for the purposes of quality control and workflow simplification. And any integration will somehow complicate the system and increase the cost, so the other key consideration is the ease of integration. Integration of electronics is generally easier compared to optics, and we are measuring intrinsic biophysical properties rather than labelling approaches; therefore, I do see great opportunities to integrate our new impedance cytometry device with a variety of cell sorting platforms.
- What do you find most challenging about your research?
The microfluidics and Lab on a chip research area is interdisciplinary in nature. My challenge is always to find the right people (e.g. students, postdoctoral fellows, collaborators) and secure sufficient resources to work on real impactful research problems.
- How do you spend your spare time?
I am trying to make a balance between work and personal life, so I mainly spend my spare time with my family members, especially my second kid is only 8 months old. I also spend some of my spare time to do physical exercise, which can help relax and leave some time for free thinking.
- Which profession would you choose if you were not a scientist?
I rarely thought about this before. Perhaps I would choose to be a doctor.
- Can you share one piece of career-related advice or wisdom with other early career scientists?
Based on my own experience, it is important to define a unique research domain based on your own expertise and the surrounding research ecosystem when early career scientists start their independent research. It is also wise to have a clear vision about what you want to achieve in the next five years.