Stuart J. Williams
University of Louisville, Louisville, KY, USA
Why is this useful?
This work demonstrates the use of a commercially-available single-sided rubber adhesive sheet to interface a rigid microfluidic chip for fluid access. This interconnect is an alternative to PDMS-based ports, whose plasma bonding characteristics may not be applicable for all rigid materials. The adhesive component of the port is incorporated with the rubber sheet; hence no additional curing time is needed. This technique provides a quick and inexpensive method to interface rigid microfluidic chips.
What do I need?
- FDA-compliant silicone rubber adhesive back, 1/16″ thick (#8991K523 through McMaster-Carr, ~US$20 for one square foot, or ~US$0.02 per square centimeter)
- A rigid microfluidic chip, with access ports prepared or predrilled. For demonstration purposes, glass used and drilled with a 0.9mm diamond rounded tip bit (diamondburs.net #HP801-009)
- A rubber punching tool. Here, a sharpened 20G needle was used (Small Parts NE-201PL-C). This is the same punching method we have used to create ports in PDMS devices, hence other similar methods may also work
- Tubing to insert into the port. Here, Tygon tubing (Small Parts TGY-010-C) was used with 0.010″ ID and 0.030″ OD
- A razor
- Tweezers (optional)
What do I do?
- Prepare and clean the surface of the rigid chip where the rubber port is to be applied.
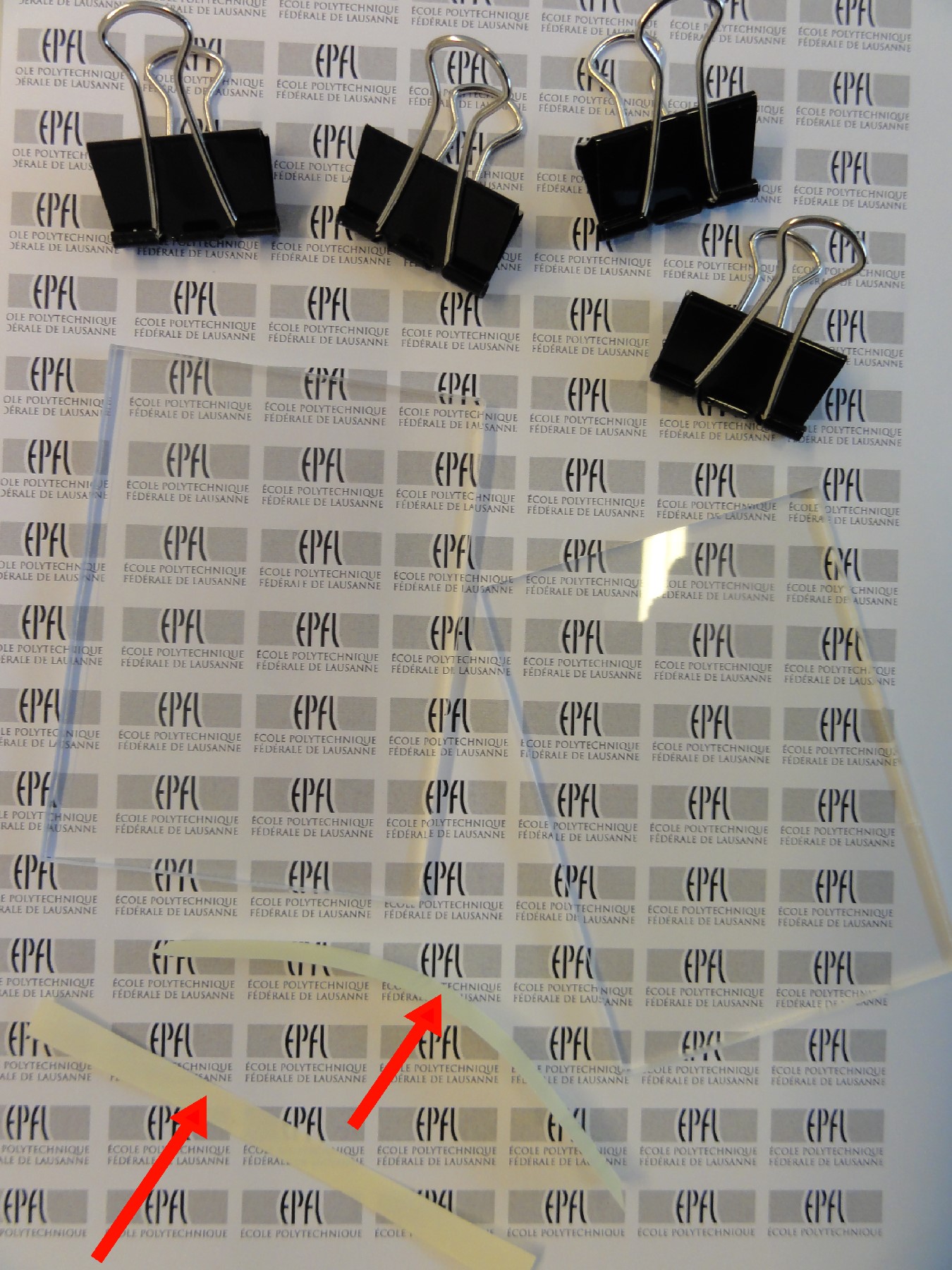
- Manually cut a small square piece of rubber from the larger sheet. This is the size of your port and should be larger than the hole on the chip. For example, a 4mm x 4mm piece was cut for a 0.9mm diameter drilled hole.
- Sharpen the 20G needle with a metal file or similar tool (Fig. 1). This provides easier punching of the sheet itself.
- Place the small rubber piece, adhesive side down, on a flat, flexible surface. We recommend using a larger excess rubber piece as a punching surface. Using the sharpened needle, manually punch through the small rubber piece. Be sure to punch through the piece completely, including the paper backing. If done successfully, the punched port will contain the bulk rubber, adhesive and backing (Fig. 2). There will not be any adhesive around the immediate vicinity of the punched hole (Fig. 3).
- Remove the paper backing on the port, exposing the adhesive.
- Align the punched hole with the microfluidic chip. Manually apply pressure to adhere the port against the chip. Note: Alignment through a transparent chip can be accomplished through visual observation through the back side of the chip. Alignment with opaque chips can be done by using a smaller gauge needle (e.g. 30G) as an alignment guide through the punched hole with the chip access hole.
- The port is ready to be used. Insert the tubing into the port with tweezers.
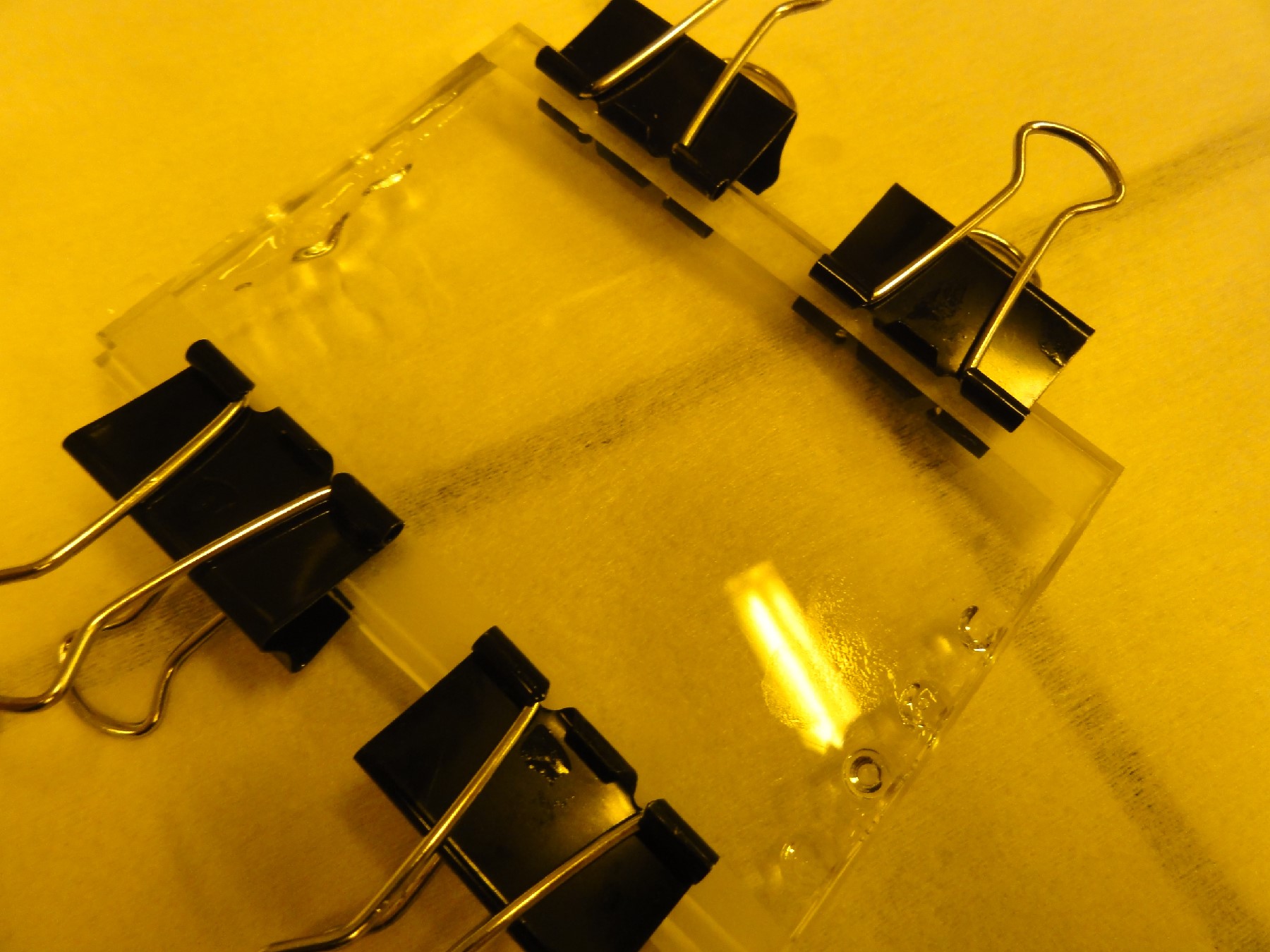
- A completed chip is shown in Fig. 4.

Fig. 1: Magnification of an original and sharpened 20G needle.

Fig. 2: Magnification of a plug after being punched from the rubber adhesive sheet. The punching process simultaneously removes the rubber bulk, adhesive, and backing.

Fig. 3: Magnification of a punched hole through the rubber sheet (bottom view). There is an absence of adhesive around the punched hole, preventing potential clogging of the port.

Fig. 4: Image of a completed chip that has two rubber microfluidic ports.
What else should I know?
Pressure limitations using this technique have not been quantified. However, no leaks were observed using manual injection methods, even after multiple uses.
Alternatively, pipette tips, syringe needles, or other items can be inserted into the rubber port for direct fluid injection.
Other adhesive rubber pads and thicknesses may work. However, other single-sided adhesive rubber pads were tested (Buna-N Rubber #8635K262, Neoprene #8583K162, Vinyl Rubber #8513K32, EPDM Rubber #8610K91, and Butyl Rubber #8609K12) and FDA Silicone performed the best overall in terms of ease of punching, bonding strength, and absence of adhesive clogs.