The 2015 Nobel Prize in Chemistry was jointly awarded to Tomas Lindahl, former director of Cancer Research UK’s Clare Hall Laboratories, Paul Modrich from Duke University in the US and Aziz Sancar from the University of North Carolina in the US for their “mechanistic studies of DNA repair”.

Tomas Lindahl, Paul Modrich and Aziz Sancar © Inserm-P. Latron, Mary Schwalm/AP/Press Association, Max Englund/UNC School of Medicine.
Tomas Lindahl’s research pieced together a molecular image of how base excision repairs DNA when a base of a nucleotide is damaged and subsequently managed to recreate the human repair process in vitro. The mechanism known as nucleotide excision repair, which excises damage from UV and carcinogenic substances, was then mapped by Aziz Sancar – the molecular details of this process changed the entire research field. Paul Modrich also studied the human version of the repair system. His work focused on DNA mismatch repair, a natural process which corrects mismatches that occur when DNA is copied during cell division.
The research carried out by the three 2015 Nobel Laureates in Chemistry has not only revolutionised our knowledge of how we function but also lead to the development of life – saving treatments. To celebrate these remarkable achievements, we are delighted to present a collection of recent Chemical Communications, Chemical Science and Chemical Society Reviews articles on DNA repair, FREE to read until 1 December 2015!
We invite you to submit your best research related to DNA repair mechanisms to Chemical Communications, Chemical Science and Chemical Society Reviews!
Reviews
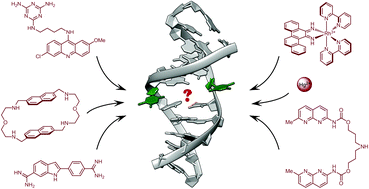
Finding needles in a basestack: recognition of mismatched base pairs in DNA by small molecules
Anton Granzhan, Naoko Kotera and Marie-Paule Teulade-Fichou
Chem. Soc. Rev., 2014, 43, 3630-3665
DOI: 10.1039/C3CS60455
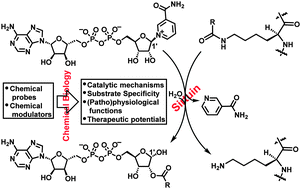
The chemical biology of sirtuins
Bing Chen, Wenwen Zang, Juan Wang, Yajun Huang, Yanhua He, Lingling Yan, Jiajia Liu and Weiping Zheng
Chem. Soc. Rev., 2015, 44, 5246-5264
DOI: 10.1039/C4CS00373J
Luminescent oligonucleotide-based detection of enzymes involved with DNA repair
Chung-Hang Leung, Hai-Jing Zhong, Hong-Zhang He, Lihua Lu, Daniel Shiu-Hin Chan and Dik-Lung Ma
Chem. Sci., 2013, 4, 3781-3795
DOI: 10.1039/C3SC51228B
Research articles
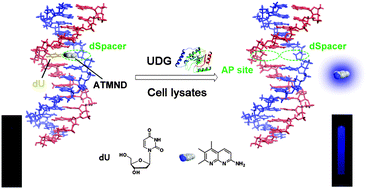
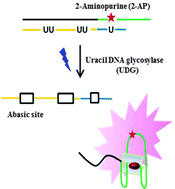
A label-free and sensitive fluorescent method for the detection of uracil-DNA glycosylase activity
Jing Tao, Panshu Song, Yusuke Sato, Seiichi Nishizawa, Norio Teramae, Aijun Tong and Yu Xiang
Chem. Commun., 2015, 51, 929-932
DOI: 10.1039/C4CC06170E
DNA-mediated supercharged fluorescent protein/graphene oxide interaction for label-free fluorescence assay of base excision repair enzyme activity
Zhen Wang, Yong Li, Lijun Li, Daiqi Li, Yan Huang, Zhou Nie and Shouzhuo Yao
Chem. Commun., 2015, 51, 13373-13376
DOI: 10.1039/C5CC04759E
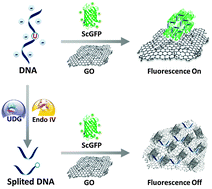
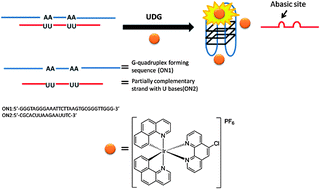
A fluorescent G-quadruplex probe for the assay of base excision repair enzyme activity
Chang Yeol Lee, Ki Soo Park and Hyun Gyu Park
Chem. Commun., 2015, 51, 13744-13747
DOI: 10.1039/C5CC05010C
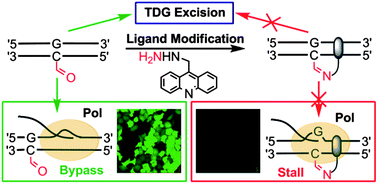
A chemical probe targets DNA 5-formylcytosine sites and inhibits TDG excision, polymerases bypass, and gene expression
Liang Xu, Ying-Chu Chen, Satoshi Nakajima, Jenny Chong, Lanfeng Wang, Li Lan, Chao Zhang and Dong Wang
Chem. Sci., 2014, 5, 567-574
DOI: 10.1039/C3SC51849C
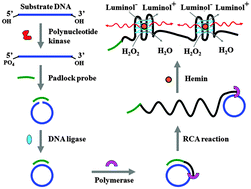
Sensitive detection of polynucleotide kinase using rolling circle amplification-induced chemiluminescence
Wei Tang, Guichi Zhu and Chun-yang Zhang
Chem. Commun., 2014, 50, 4733-4735
DOI: 10.1039/C4CC00256C
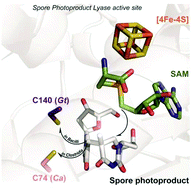
Rescuing DNA repair activity by rewiring the H-atom transfer pathway in the radical SAM enzyme, spore photoproduct lyase
Alhosna Benjdia, Korbinian Heil, Andreas Winkler, Thomas Carell and Ilme Schlichting
Chem. Commun., 2014, 50, 14201-14204
DOI: 10.1039/C4CC05158K
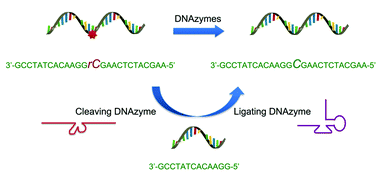
Expanding DNAzyme functionality through enzyme cascades with applications in single nucleotide repair and tunable DNA-directedassembly of nanomaterials
Yu Xiang, Zidong Wang, Hang Xing and Yi Lu
Chem. Sci., 2013, 4, 398-404
DOI: 10.1039/C2SC20763J
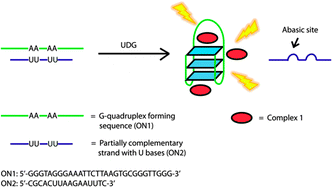
Detection of base excision repair enzyme activity using a luminescent G-quadruplex selective switch-on probe
Ka-Ho Leung, Hong-Zhang He, Victor Pui-Yan Ma, Hai-Jing Zhong, Daniel Shiu-Hin Chan, Jun Zhou, Jean-Louis Mergny, Chung-Hang Leung and Dik-Lung Ma
Chem. Commun., 2013, 49, 5630-5632
DOI: 10.1039/C3CC41129J
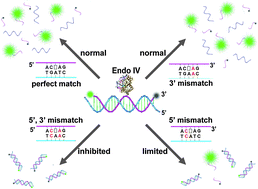
Endonuclease IV discriminates mismatches next to the apurinic/apyrimidinic site in DNA strands: constructing DNA sensing platforms with extremely high selectivity
Xianjin Xiao, Yang Liu and Meiping Zhao
Chem. Commun., 2013, 49, 2819-2821
DOI: 10.1039/C3CC40902C
Also of interest: Find out more about the three Chemistry Nobel Laureates and their research.
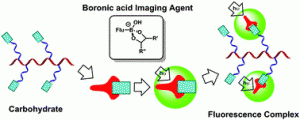 The well-known and strong affinity of boronic acids for carbohydrates offers a convenient means of detecting commonly expressed markers in diseases including some cancers, as well as Alzheimer’s, autoimmune, and heart diseases. As such, the attachment of this relatively simple chemical moiety to fluorescent small molecular, polymeric or benzoxaborale-based probes offers a diagnostic tool that is able to detect, monitor, and aid in the personalised treatment of such significant and life-changing diseases.
The well-known and strong affinity of boronic acids for carbohydrates offers a convenient means of detecting commonly expressed markers in diseases including some cancers, as well as Alzheimer’s, autoimmune, and heart diseases. As such, the attachment of this relatively simple chemical moiety to fluorescent small molecular, polymeric or benzoxaborale-based probes offers a diagnostic tool that is able to detect, monitor, and aid in the personalised treatment of such significant and life-changing diseases. Anthea Blackburn is a guest Web Writer for Chemical Communications. Anthea hails from New Zealand, carried out her graduate studies in mechanostereochemistry under the guidance of Prof. Fraser Stoddart in the US, and has recently relocated to live in London. She is a recent addition to the Econic Technologies team, where she is working on the development of new catalysts for the environmentally beneficial preparation of polycarbonates from CO2.
Anthea Blackburn is a guest Web Writer for Chemical Communications. Anthea hails from New Zealand, carried out her graduate studies in mechanostereochemistry under the guidance of Prof. Fraser Stoddart in the US, and has recently relocated to live in London. She is a recent addition to the Econic Technologies team, where she is working on the development of new catalysts for the environmentally beneficial preparation of polycarbonates from CO2.
























 As Alzheimer’s disease advances, symptoms can include confusion, irritability and aggression, and long-term memory loss © Shutterstock
As Alzheimer’s disease advances, symptoms can include confusion, irritability and aggression, and long-term memory loss © Shutterstock![spinksbanner2_tcm18-223978[1]](https://blogs.rsc.org/md/files/2012/11/spinksbanner2_tcm18-2239781.jpg)