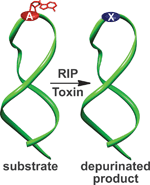
A class of protein toxins exists with an extremely apt acronym, RIP. This stands for Ribosome Inactivating Proteins but may as well stand for “Rest in Peace” when applied to cellular RNA. These proteins are known to attack the link between a purine base (adenine or guanosine) and its sugar causing depurination. This results in the formation of abasic sites (bases that are neither pyrimidines or purines) which in turn, causes the ribosome containing the sequence to have a lower affinity to elongation factors that are crucial for protein synthesis, ultimately leading to cell death.
Methods exist to detect RIPs such as ELISAs and antibody-based immunoassays or by monitoring the specific depurination activity through fluorescence, radiolabelling and immunoaffinity chromatography, amongst others. These techniques require sophisticated or elaborate set-ups, limiting the potential for high throughput (HTP) screening in a bid to discover potential inhibitors of these destructive toxins.
Seergazhi Srivatsan and co-workers have produced a label-free fluorescence hybridisation assay to detect the depurination activity of saporin, a RIP toxin, using a fluorescence ligand that specifically binds to the cytosine opposite an abasic site. If depurination takes place, then the ligand can bind and its fluorescence intensity will be quenched. This technique allows for a plethora of information to be obtained about the depurination activity of saporin as well as many other RIP toxins. This opens the door to using high throughput screening to find inhibitors of such toxins.
Download the article to read more…
Also of interest: Overcoming obstacles in labelling RNA
Posted on behalf of Sarah Brown, web science writer for ChemComm.